Calag Archive
Calag Archive
Buffers between grazing sheep and leafy crops augment food safety
Publication Information
California Agriculture 67(2):104-109. https://doi.org/10.3733/ca.v067n02p104
Published online April 01, 2013
PDF | Citation | Permissions
Abstract
The presence of livestock in or near fresh-market vegetable fields has raised concerns about the potential for contaminating produce with pathogenic bacteria. To develop buffer zones for grazing near production of leafy greens, we assessed the prevalence of Escherichia coli O157:H7 and Salmonella species in sheep that were grazed on alfalfa fields during the winter in California's Imperial Valley. We found E. coli O157:H7 in 1.8% of fecal samples and 0.4% of soil samples, and Salmonella in 0.8% of fecal samples and 0.4% of soil samples. Our results indicate that sheep grazing on alfalfa in the Imperial Valley have a low prevalence of these pathogens in their feces and that these bacteria are rarely found in soil from fields with grazing sheep. The California Leafy Green Products Handler Marketing Agreement guideline of 30 feet between grazing lands or domestic animals and the crop edge is adequate to minimize potential contamination of nearby crops.
Full text
California ranks second in the nation for sheep production, yielding 325,000 lambs, more than 3 million pounds of wool, and $50 million annually (CDFA 2007). The sheep industry relies heavily on grazing crop, vineyard and orchard fields throughout California as a source of inexpensive feed. In addition, ruminants play an important role in sustainable agricultural systems. Sheep are particularly useful in converting vast renewable resources including rangeland and pasture forage as well as crop residues into edible food (Oltjen and Beckett 1996). Moreover, the manure produced by the sheep serves as an organic fertilizer that improves soil structure and contributes to plant nutrition.
Wherever sheep graze, they always leave something behind — as on this alfalfa field, where their droppings become fertilizer for subsequent crops. But if the animals range too close to a field where food crops are grown, there's a potential for contamination with disease-causing bacteria.
The sheep production system employed in Imperial County, California, involves intensive grazing for short time periods. Bands of approximately 1,500 sheep are typically left in a 40-acre field until the forage is grazed close, which usually takes 4 to 7 days. Then the sheep are moved to another field. If the next field is located only 2 to 3 miles away, this is often accomplished by herding the flock along public roads.
Sheep grazing is seasonal in the Imperial Valley. Typically, sheep arrive from throughout the western United States in October and rotate between fields of alfalfa or bermudagrass until approximately mid- to late March, when they are sent to a feedlot or directly to slaughter. Market prices for sheep and alfalfa and other factors (such as transport cost) affect the use of this grazing system.
Rotating vegetables and alfalfa
The Imperial Valley has long been recognized as the “winter salad bowl” for the United States. With over 100,000 acres of fresh-market vegetable production that is valued at $500 million and distributed nationwide, the valley's agricultural industry has a tremendous impact on the local economy as well as on the nationwide food supply (CDFA 2007). To produce fresh-market vegetables successfully, growers must rotate these crops with others that provide a suitable economic return while reducing pest pressure in the subsequent vegetable crop. Alfalfa is the standard rotation with vegetable crops in Imperial County.
Leafy greens and bacteria
Integrating crop and animal agriculture, however, can have detrimental consequences. Crop contamination with Escherichia coli O157:H7 (which in humans can cause severe acute hemorrhagic diarrhea, and in a small proportion of cases can lead to hemolytic uremic syndrome and possibly kidney failure) has been documented through application of raw manure, use of irrigation water contaminated with animal or human feces (Solomon et al. 2002) and deposition of feces by livestock and wild animals (Cieslak et al. 1993; Jay et al. 2007). Recent outbreaks of human disease caused by E. coli O157:H7 in California have been associated with consumption of raw spinach (CDC 2006; Jay et al. 2007) and lettuce (Brandl 2008; Cooley et al. 2007). These have raised concerns that sheep and other ruminants may elevate levels of pathogens within the soil, which then have the potential of being transmitted to produce fields via aerosols (CDC 2006; Cieslak et al. 1993; Cooley et al. 2007; Jay et al. 2007).
The California Leafy Green Products Handler Marketing Agreement (LGMA) of January 2012 lists sheep as one of the five mammalian species that are “animals of significant risk” and states that any intrusion by such animals requires a detailed food safety assessment prior to harvest (LGMA 2012). Buffer zones between the crop production fields and livestock operations are important in order to prevent the potential transmission of pathogens from animals to crops.
Due to food safety concerns, essentially all of California's leafy greens are produced and marketed under the California Leafy Green Products Handler Marketing Agreement (LGMA 2012). The participating companies have committed themselves to selling products grown in compliance with LGMA food safety practices, including having a complete food safety compliance program, performing environmental assessments, testing and record-keeping for water use and soil amendments, to name a few.
The LGMA recognizes the need for further research to validate or adjust these guidelines based on scientific evidence. One area identified by LGMA as needing additional research relates to setback distances, or buffer zones, between livestock and crops. The LGMA suggests a buffer of 400 feet between the edge of a crop and a concentrated animal-feeding operation, and a buffer of 30 feet for grazing lands or domestic animals. However, they also acknowledge a lack of science on which to base this recommendation (LGMA 2012). There is a paucity of information related to appropriate combinations of time and distance between livestock operations and crop systems, particularly with regard to how long pathogens survive in animal feces, soil and aerosols, and how pathogens move via wind, water or flies.
Leafy greens and sheep
While considerable attention has been paid to the prevalence of foodborne pathogens in cattle, less is known about the epidemiology of E. coli O157:H7 in grazing sheep. Much as in cattle, prevalence of this organism in sheep varies considerably, with levels as low as 0.2% reported in some studies (Battisti et al. 2006) and as high as 68% in others (Sidjabat-Tambunan and Bensink 1997). There are approximately 650,000 sheep and lambs in California and as many as 150,000 in the Imperial Valley seasonally, making it essential to understand the ecology of important human pathogens associated with sheep. Therefore, the primary objective of this research project was to develop data that can be used to accurately define buffer zones appropriate for grazing of sheep near production of leafy greens.
Measuring fecal bacteria
Fecal and soil samples were collected from alfalfa fields where bands of sheep were grazing or had recently grazed. These bands consisted of 1,200 to 1,800 head of approximately 6- to 10-month-old lambs from numerous locations throughout the western United States. Typically, a 160-acre block of alfalfa is broken into four 40-acre plots, and each plot is grazed for approximately 7 days. These blocks are separated from other crops by either an irrigation ditch or a roadway. Each field that was sampled for this study was stocked with sheep only once during the winter grazing season. For each collection, 40 samples of fresh feces (minimum 0.35 ounce or 10 grams) and 40 samples of soil (minimum 0.35 ounce or 10 grams) were placed into individual containers, which were immediately placed on ice. Samples were shipped overnight by courier and processed within 24 hours of collection. Data were recorded and related to management of the sheep and the alfalfa field, including number of days sheep were present on the current field, age and source of sheep, and number of days since last irrigation event. Most bands of sheep were sampled once, but four bands were sampled twice, and two bands were sampled three times.
Placed downwind from grazing livestock, a portable microbial air monitoring station keeps track of airborne bacteria.
Standard microbiological techniques were used to enumerate commensal E. coli (those generally regarded as non-pathogenic) (APHA 2012) and to identify two potential pathogens, E. coli O157:H7 (Kilonzo et al. 2011) and Salmonella species (US EPA 2011). We determined mean coliform bacteria concentration in feces and soil by dispersing 0.035 ounce (1.0 gram) of feces or soil in 1.32 fluid ounces (39 milliliters) of phosphate buffered solution (PBS) using a rotational mixer for 5 minutes. The feces- or soil-PBS solution was then serially diluted (102, 103, 104, 105, 106). The E. coli concentration in the diluted feces- or soil-PBS solution was determined by direct membrane filtration and culturing onto CHROMagar EC (Chromagar Microbiology, Paris, France) at 112.1°F (44.5°C) for 24 hours (APHA 2012).
Above, one side of an Imperial Valley road is a leafy green vegetable crop; the other, an alfalfa field. As many as 150,000 sheep graze on Imperial Valley alfalfa from October to March. During this period, sheep are moved to a new field every 4 to 7 days, and at times herded along public roads.
Fecal and soil samples were enriched for Salmonella species using US EPA Method 1682 (US EPA 2011). Escherichia coli O157 samples were enriched in tryptic soy broth (TSB), exposed to an immunomagnetic separation step and then cultured on cefixime potassium tellurite sorbitol MacConkey (CT SMAC) and Rainbow agar containing novobiocin and tellurite (NT Rainbow) as previously described (Cooley et al. 2007; Kilonzo et al. 2011). E. coli O157:H7 colonies identified were further analyzed by real-time PCR (RT-PCR) to detect presence of virulence genes. Pulsed-field gel electrophoresis was performed on E. coli O157:H7 isolates with the standard PulseNet procedure by using XbaI restriction enzyme (Kilonzo et al. 2011; Ribot et al. 2006).
Air samples were collected from the edge of the field where the sheep were grazing. The prevailing wind direction was used to determine which side of the field was sampled. Samples were collected from the downwind side of the field in duplicate or triplicate at the following distances from the field edge: 6.6, 16.4, 32.8, 65.6, 164.0 and 328.1 feet (2, 5, 10, 20, 50 and 100 meters). A sample was also obtained from an upwind location to serve as a control. The microbial air monitoring system MAS-100Eco (Merck) was used to test total levels of all forms of bacteria. The MAS-100 aspirates air at the rate of 26.4 gallons (100 liters) of air per minute, and after initial tests we determined that a sampling time of 10 minutes was appropriate given the low concentrations of bacteria in the air. Specific agar (Chromocult) was used to enumerate colonies, and the number was converted to colony-forming units per cubic meter of air (Chinivasagam et al. 2009). Air samples were obtained at the same time and on the same day as the fecal and soil collections. Air samples were collected on five additional occasions as well.
Meteorological data (wind speed, temperature, relative humidity, rainfall) were recovered daily from the closest California Irrigation Management Information System (CIMIS) weather station (CIMIS 2012). Data were entered into, and graphs were prepared from, a commercially available spreadsheet program (Excel 2010, Microsoft Corporation, Redmond, WA). Statistical analysis (chi-square tests for homogeneity, one-way analysis of variance) was performed using a software program (Egret for Windows, version 2.0.1, Cytel Software Corporation, Cambridge, MA).
Pathogenic bacteria from sheep
Since sheep are present only seasonally in the Imperial Valley, samples were collected from February 2011 to March 2011 and again from October 2011 to December 2011. Total precipitation during these two time periods was 1.82 inches (4.62 centimeters); average air temperature was 57.5°F (14.2°C) and average wind speed was 4.3 mph (6.9 kilometers per hour). Ten-year averages during the period of October to March are 1.1 inches (2.9 centimeters) rainfall, 61.1°F (16.2°C) average air temperature and 4.5 mph (7.3 kilometers per hour) average wind speed (CIMIS 2012).
A total of 1,440 samples (720 fecal and 720 soil) were collected throughout the project.
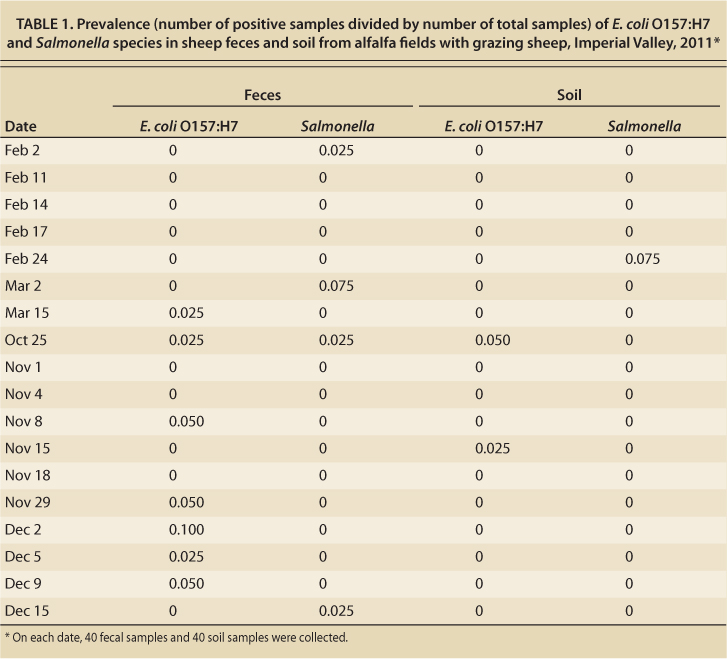
E. coli O157:H7.These pathogenic coliform bacteria were found in 13 of the fecal samples (1.8%) and in 3 of the soil samples (0.4%) (table 1). E. coli O157:H7 was found in feces in 7 of 18 sample collections, and in soil in 2 of 18 sample collections. The highest prevalence of E. coli O157:H7 in feces at any one sample collection was 10% (4 positive out of 40 samples).
TABLE 1. Prevalence (number of positive samples divided by number of total samples) of E. coli O157:H7 and Salmonella species in sheep feces and soil from alfalfa fields with grazing sheep, Imperial Valley, 2011
There were no statistically significant differences in the proportion of positive samples on any of the collection dates; however, there were more positive fecal samples collected from October to December (12 of 440) than from January to March (1 of 280) (p value = 0.02). No significant associations between prevalence and management factors such as duration of grazing, irrigation events or source of sheep were detected. There was also no association between duration of sheep grazing and presence of bacteria in the soil.
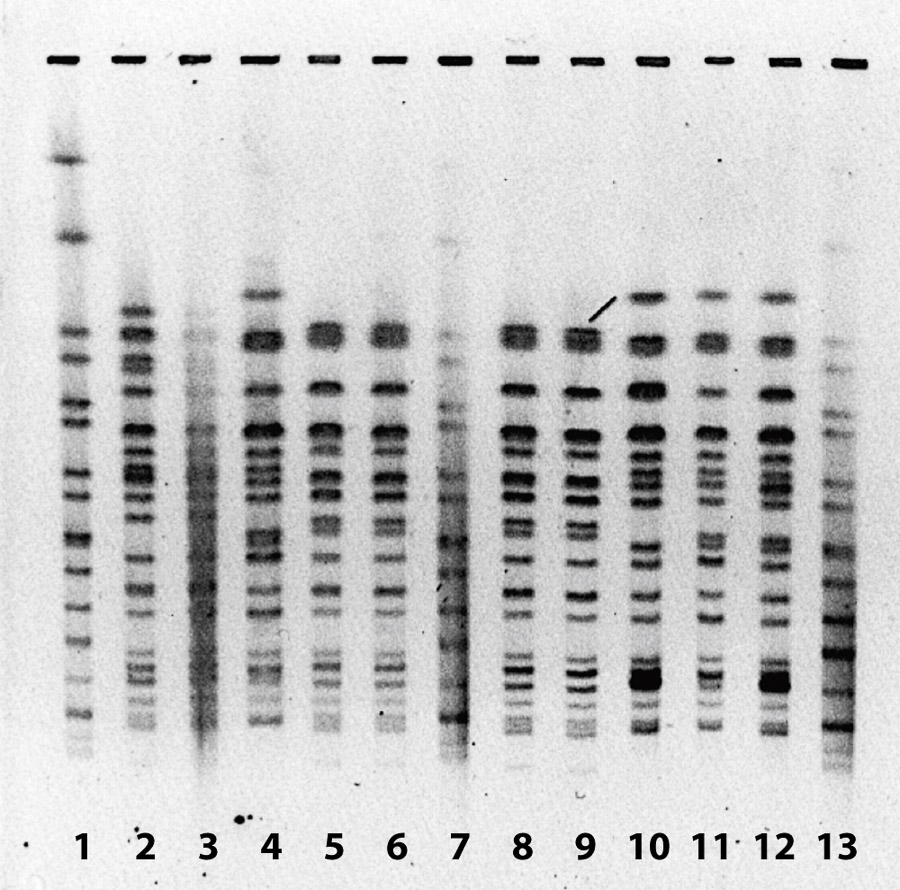
Pulsed-field gel electrophoresis (PFGE) was performed to determine genetic relatedness of isolates on several of the recovered E. coli O157:H7 samples (fig. 1). In general, isolates from the same date and same group of sheep shared a PFGE pattern, while isolates from other groups of sheep sampled on different dates had unique patterns. This indicates that different bands of sheep likely carry unique strains of E. coli O157:H7, a factor that could be useful in determining bacterial sources in the event of a disease outbreak.
Fig. 1. Pulsed-field gel electrophoresis of E. coli O157:H7 isolates from sheep feces and soil with grazing sheep, Imperial Valley, 2011. Lanes 1, 7 and 13 are control isolates. Lanes 2 and 3 are from Oct. 25, 2011, lane 4 from Nov. 8, 2011, lanes 5, 6, 8, and 9 from Dec. 2, 2011, lane 10 from Dec. 5, 2011, and lanes 11 and 12 from Dec. 9, 2011. Samples collected from different days have different patterns, indicating the presence of different E. coli O157:H7 genotypes.
Salmonella species.
These potentially pathogenic bacteria were detected in 6 fecal samples (0.8%) and 3 soil samples (0.4%). All positive soil samples were obtained on the same sampling date, while positive fecal samples were obtained from three sampling dates. Interestingly, a significant precipitation event (over 2.5 centimeters, 1 inch, within a 2-day period) occurred a few days before the Salmonella positive soil samples were collected (table 1). We speculate that the additional moisture likely affected the ability of the bacteria to survive and therefore be recovered from the soil.
In the corner of an alfalfa field, grazing sheep cluster near a water trough. Investigators analyzed soil and fecal samples from fields intensively grazed by bands of sheep; each consisted of 1,200 to 1,800 head of approximately 6- to 10-month-old lambs, from various locations in the western United States.
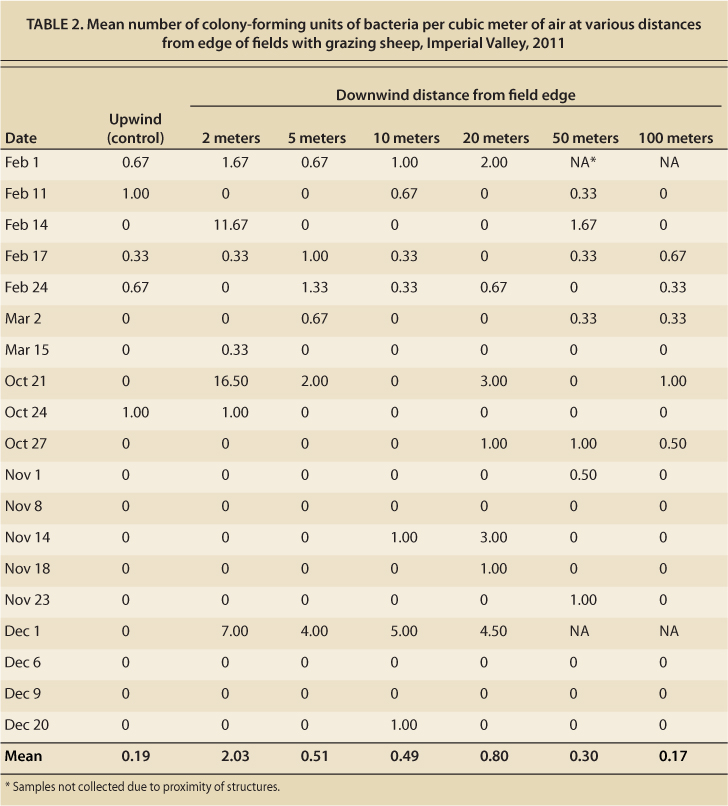
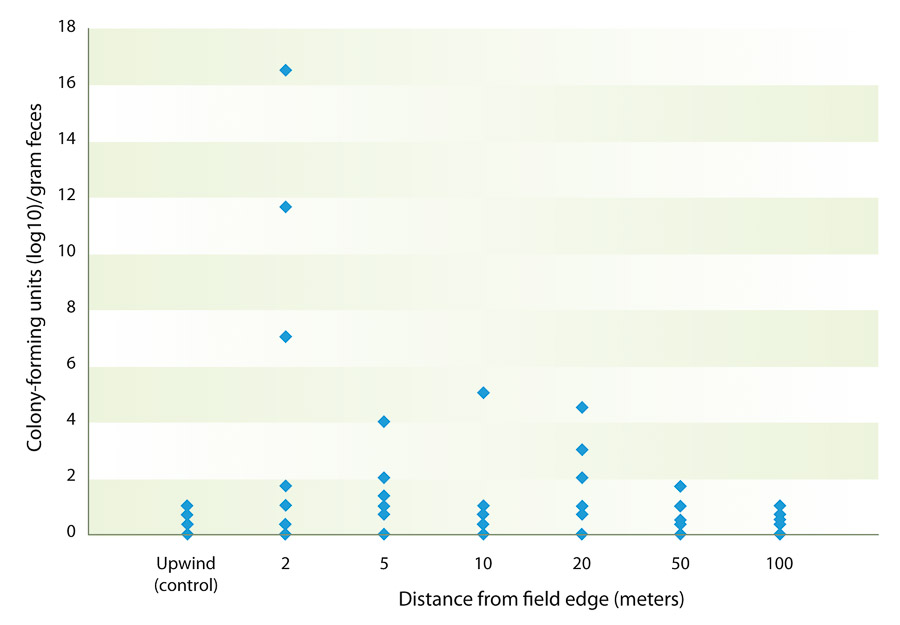
The mean commensal E. coli and coliform bacteria concentration in feces and soil was also measured. The overall mean coliform count from feces over the entire project was 1.05 × 107 colony-forming units per gram of feces, while mean commensal bacteria count from soil was 3.5 × 103 colony-forming units per gram of soil. This compares well to the mean coliform counts of approximately 6 × 106 found by other researchers (Taylor et al. 2012). Air sampling revealed that few bacteria were being dispersed through the air. The maximum number of colony-forming units was 16.5 per cubic meter of air obtained from a sample collected on October 21, 2011. The mean number of colony-forming units was greatest at a distance of 2 meters from the alfalfa field edge; however, a one-way analysis of variance demonstrated that there was no statistically significant difference at any distance measured, nor was there significant correlation between distance and bacterial count. Using linear regression, it was determined that there were no significant correlations between fecal or soil E. coli counts and aerosol bacterial counts at any of the distances measured (table 2, fig. 2).
TABLE 2. Mean number of colony-forming units of bacteria per cubic meter of air at various distances from edge of fields with grazing sheep, Imperial Valley, 2011
Fig. 2. Mean number of colony-forming units of bacteria per cubic meter of air from samples obtained at various distances from the edge of fields with grazing sheep, Imperial Valley, 2011. Each data point shown represents a bacteria level measured on one or more sampling dates from February 1 to December 20, 2011. (Specific data by date are listed in table 2.)
Feces and soil are low risk
We detected E. coli O157:H7 in 1.8% of fecal samples and 0.4% of soil samples, and Salmonella species in 0.8% of fecal samples and 0.4% of soil samples. These results indicate that fresh sheep feces present minimal risk for leafy greens producers. Soil where sheep have been actively grazing is even lower risk. The positive soil samples were observed to be associated with recent (within 5 to 7 days) rainfall events. However, this observation requires further study, as positive soil samples were obtained in only 3 sample collections.
The finding of any human pathogens in fresh sheep feces warrants the LGMA recommendation of a food safety assessment following known intrusion by sheep. The PFGE patterns indicate that unique strains of E. coli O157:H7 exist in populations of sheep, and this information can be useful if investigations of the source of contamination of leafy greens are necessary.
Airborne transmission of bacteria was also assessed in this project. The greatest number of airborne bacteria (from all sources including feces) recovered from the edge of a field where sheep were grazing was at a distance of 2 meters (table 2, fig. 2). However, the difference compared to the number of bacteria at any other distance was not significant. We did not speciate the bacteria found. Given the low prevalence of pathogenic bacteria in feces and soil, we were not able to detect these pathogens in air samples. (Although we did not speciate bacteria from airborne samples, their gross morphology—or appearance on the agar plates—did not match that of pathogenic bacteria.) Future studies that compare the genotypes of bacteria recovered by air sampling with bacteria from the sheep would be worthwhile to improve understanding of the possibility of airborne transmission.
Above, portable electric fencing restricts movement of grazing livestock. Based on low prevalence of pathogenic bacteria in soil and fecal samples, scientists concluded that a buffer distance of 30 feet between grazing sheep and the edge of a food crop was adequate to prevent contamination.
Based on our findings, we believe that the LGMA-recommended buffer distance of 30 feet (9 meters) between grazing livestock and crops is justified, and is more than adequate to ensure protection from potential contamination.