Posts Tagged: e. coli
Science-based food safety tips
Last month, I attended ScienceWriters2014, a joint meeting of the National Association of Science Writers, Inc, and the Council for the Advancement of Science Writing, in Columbus, Ohio. Held in cooperation with The Ohio State University, the conference attracted 430 freelancers, students, editors, staff journalists, public information officers and other lovers of science and science-writing. I had applied for a public information officer travel fellowship to attend it, and was fortunate to be awarded one by the NASW, greatly facilitating my attendance.
One of the events that attendees could sign up for was lunch with a scientist at The Ohio State University, located a few miles away from the conference venue. I chose Prof. Jeffrey LeJeune, an infectious disease microbiologist and epidemiologist, because a focus of his research is food safety, one of the topics included in the UC Global Food Initiative that UC President Janet Napolitano launched on July 1.
On the day of the luncheon, a Sunday, we were driven to The Ohio State University in buses the university provided. We assembled in the lobby of the Ohio Union (it was homecoming on campus and the Columbus marathon was in progress nearby), and were soon escorted to the tables of the scientists we had picked. The university kindly (and safely!) provided lunch.
At LeJeune's table, we introduced ourselves to one another. LeJeune began his presentation to his 15 guests by rebuffing the five-second rule. According to this rule, food dropped on the ground will not become contaminated with bacteria if it is picked up within five seconds of being dropped. LeJeune said it does not work. “Eating off the floor violates all food-borne illness prevention advice,” he warned.
Perhaps because we in his audience were all science writers, he proceeded to discuss communication challenges facing scientists. He said most of the emphasis in graduate training is on making discoveries, with hardly any attention paid to communicating these discoveries in lay language to benefit the general public. Other challenges he mentioned are the information explosion we are witnessing, resulting in deaf ears turned to many scientists' voices; and language barriers between scientists and journalists that hinder effective communication.
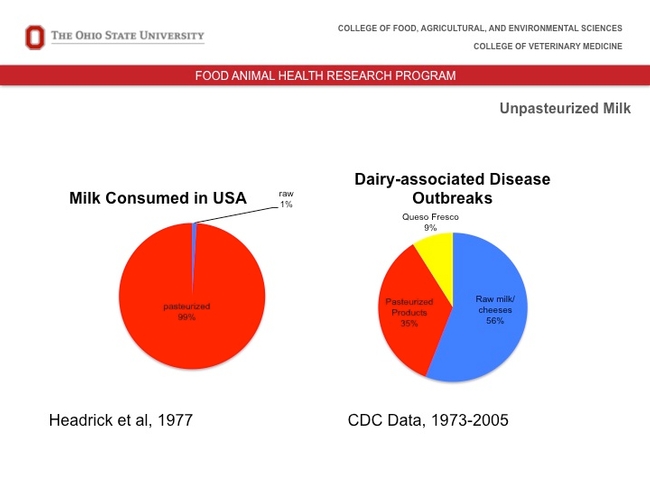
LeJeune then introduced the topic of raw milk. He said that while consuming raw milk is dangerous (CDC data for 1973-2005 shows that 56 percent of dairy-associated disease outbreaks result from raw milk/cheeses), less than one percent of milk consumed in the United States is raw.
“The pasteurization of milk was a huge benefit to the health of the human population,” he said. “Most cheeses in the U.S. are pasteurized cheese products.”
We asked him many questions. He answered them all. He explained that the U.S. has the safest food supply. Despite this, pathogens can enter the food chain through live animals, he cautioned. Further, refrigeration could be inadequate. He said about 80 percent of food and vegetable contamination occurs post-farm. His tip for what to eat when traveling: “Avoid raw or unpeeled foods. It is best to choose what is fully cooked and hot.”
LeJeune noted there is no evidence to suggest that GM foods are problematic from a food safety perspective.
“There are some concerns for sure,” he said. “But these are largely economic or political. Nutrition-wise, GM foods can be beneficial. From a food safety and nutritional standpoint, I also see no significant differences between organic produce and non-organic or regular produce. There could be, however, some environmental impacts related to the different production systems.”
More questions followed. A discussion on E. coli bacteria gathered momentum, specifically how E. coli gets infected with a virus and how, when this virus decides to leave E. coli, it releases Shiga toxins, which, in turn, damage cells lining the kidney.
We were so engrossed in the discussion that it came as a surprise when one of the organizers of the luncheon strode into the room to inform us that our hour with the scientist was up and that the bus that had transported us to The Ohio State University was about to leave.
As we rose hastily from our chairs we thanked LeJeune for his presentation, which was clear and to the point – qualities all science (and other) writers appreciate. We know he had other topics to discuss with us: Can I cook my Jack-o'-Lantern after Halloween? (The answer is “Not if it sits out for more than two hours.”) And are raw diets for dogs a public health concern for humans? (The answer is “Your dog is more likely to have Salmonella if it is eating raw food.)
Although we didn't get to these topics, he left us with ample useful information about food safety. On the ride back to the conference, the bus was loud with conversation from the various lunch groups – what had been learned, how best it could be communicated, and how each one of us had made a new friend at the university.
Foodborne illnesses and the 100K Genome Project
An ambitious effort to sequence the genomes of 100,000 infectious microorganisms and speed diagnosis of foodborne illnesses has been launched by the University of California, Davis, Agilent Technologies, and the U.S. Food and Drug Administration.
Bart Weimer, professor in the UC Davis School of Veterinary Medicine, serves as director of the 100K Genome Project and co-director of the recently established BGI@UC Davis facility, where the sequencing will be done. Other collaborators include the U.S. Centers for Disease Control and Prevention and the U.S. Department of Agriculture.
The new five-year microbial pathogen project focuses on making the food supply safer for consumers. The group will build a free, public database including sequence information for each pathogen's genome — the complete collection of its hereditary information. The database will contain the genomes of important foodborne pathogens including Salmonella, Listeria, and E. coli, as well as the most common foodborne and waterborne viruses that sicken people and animals.
The project will provide a roadmap for developing tests to identify pathogens and help trace their origins more quickly. The new genome database also will enable scientists to make discoveries that can be used to develop new methods for controlling disease-causing bacteria in the food chain.
"This landmark project will revolutionize our basic understanding of these disease-causing microorganisms," said Harris Lewin, vice chancellor for research at UC Davis.
The sequencing project is critically important for tackling the continuing outbreaks of often-deadly foodborne diseases around the world. In the United States alone, foodborne diseases annually sicken 48 million people and kill 3,000, according to the CDC.
"The lack of information about food-related bacterial genomes is hindering the research community's ability to improve the safety and security of the world food supply," Weimer said. "The data provided by the 100K Genome Project will make diagnostic tests quicker, more reliable, more accurate and more cost-effective."
"We see this project as a way to improve quality of life for a great many people, while minimizing a major business risk for food producers and distributors," said Mike McMullen, president of Agilent’s Chemical Analysis Group.
A consumer-focused article about the project is available on the FDA website.
(This article was condensed from a UC Davis news release. Read the full press release and watch a video of Bart Weimer giving an overview of the project.)
Scientists target E. coli from farm to fork
The University of California, Davis, is participating in a large-scale research effort aimed at preventing potentially fatal illnesses linked to E. coli bacteria.
The U.S. Department of Agriculture is funding the $25 million, coast-to-coast project, to which UC Davis is providing expertise in livestock health, foodborne disease and consumer food marketing.
The project, announced Jan. 23 by the USDA, aims to reduce the occurrence of and public health risks associated with Shiga toxin-producing E. coli. The research effort is led by the University of Nebraska, Lincoln.
UC Davis researchers collaborating in the project include James Cullor, a professor in the School of Veterinary Medicine; Christine Bruhn, a food science marketing specialist and director of the Center for Consumer Research; and Terry Lehenbauer, director, and Sharif Aly, assistant professor, both of the Veterinary Medicine Teaching and Research Center in Tulare.
Cullor and his colleagues in the veterinary school's Dairy Food Safety Laboratory -- in Davis and Tulare -- will conduct research aimed at reducing the microbial counts on cattle hides during processing, looking for ecologically responsible methods for enhancing food safety. They also will test radiofrequency technologies, which use electrical currents oscillating at specific frequencies to inactivate E. coli on beef carcasses during processing.
Bruhn will collaborate with North Carolina State University and Kansas State University to reduce health risks associated with undercooked hamburgers. The researchers will encourage television food programs to include safe food-handling practices and messages.
In addition, Bruhn will work with health care professionals to raise the number of food-handling messages directed toward consumers who are at increased risk for foodborne illness, especially children and people with diabetes. She also will investigate consumer interest in the use of irradiation or high-pressure technologies to enhance the safety of ground meat.
Lehenbauer, Aly and their colleagues at the Veterinary Medicine Teaching and Research Center will participate in animal research needed for understanding the epidemiology and ecology of non-Shiga toxin-producing E. coli, after information from preliminary studies is used to develop the scientific protocols for these animal-sampling projects. The research team will focus on dairy cattle, including male Holstein cattle that are being raised for beef production
Other participants are the University of Delaware, New Mexico State University, Texas A&M, Virginia Tech, the University of Arkansas, the USDA's Agricultural Research Service, and a consortium of government, academic and industry scientists and food safety professionals.
Research helps ensure safety of leafy greens
Four years ago, a multi-state outbreak of E. coli O157:H7 in fresh baby spinach gripped the nation. Nearly 200 people in 26 states came down with the disease. Two elderly women and a 2-year-old boy died.
The outbreak was also devastating for the industry. The contaminated spinach was traced back to Central California, where growers produce 80 percent of the nation’s leafy greens. Scientists, farmers and regulators worked together to restore public confidence in products that are widely considered part of a healthy diet. Regulators and farmers created the California Leafy Green Marketing Agreement to establish a culture of food safety on leafy greens farms and researchers worked to close gaps in the body of scientific knowledge about the sources of E. coli O157:H7 in the region.
In 2006, UC and USDA researchers were already designing a four-year study of the possible sources of E. coli O157:H7 near Central California fresh produce fields when the high-profile spinach outbreak occurred. This month, data collection from rangeland and farmland, steams and irrigation canals comes to a close. The team of scientists is now analyzing the data to reach conclusions that will help prevent future food contamination.
Preliminary results reflect a diversity of E. coli O157:H7 carriers near Central Coast farms, according to Edward (Rob) Atwill, a UC Davis School of Veterinary Medicine specialist in waterborne infectious diseases and co-principal investigator of the study. Early on, free-ranging feral swine were implicated as carriers of the deadly bacteria, but it wasn’t known whether there were other sources in the environment. The researchers collected 1,233 samples of wild and feral animal scat from 38 Central Coast cattle ranches and leafy greens farms that were adjacent to riparian, annual grassland and oak woodland habitat. Eighteen of the samples were found to contain E. coli O157:H7.
The scientists found the bacteria in
- 3 of 60 brown-headed cowbirds
- 5 of 93 American crows
- 2 of 95 coyotes
- 1 of 72 deer mice
- 10 of 200 feral swine
E. coli O157:H7 was not found in scat samples from deer, opossums, raccoons, skunks, ground squirrels, or other bird and mouse species.
“Our goal over the next nine months is to finish analyzing this very large and comprehensive dataset and to identify various good agricultural practices that reduce the risk of foodborne pathogens for the produce industry,” Atwill said.

Research helps prevent contamination of fresh leafy greens.
E. coli outbreak on lettuce parallels recent
UC research development
Although the outbreak earlier this month of E. coli O145 in shredded Romaine lettuce hasn’t touched California consumers and local retailers, it is impacting the industry. Once again, the safety of pre-washed and cut leafy vegetables are in headlines, raising the fears of consumers and producers alike.
The Centers for Disease Control have confirmed 23 illnesses and 7 probable illnesses in New York, Michigan, Ohio and Tennessee from the contaminated lettuce. The traceback investigation is stretching west to Yuma, Ariz., where the lettuce may have been grown. So far, the source of the contamination has not been identified.
A source of confusion and alarm, however, is the contaminant’s O145 designation. For the past 30 years, the majority of the E. coli illness outbreaks in America were serotype O157:H7. UC Davis Cooperative Extension produce safety specialist Trevor Suslow said E. coli O145 doesn’t come completely out of the blue. O145 is recognized in its association with food in other countries and more often with animal products, not produce.
Suslow and his staff were well aware of E. coli O145 prior to this month’s outbreak and included it in their recent research aimed at development of a new method for detecting a broad range of the most dangerous E. coli bacteria in produce and produce production environments.
The new test was not designed to identify a particular strain of E. coli, but to determine whether a pathogenic/toxigenic strain of E. coli – such as O145 – is present. If the test comes up positive, a secondary test can be conducted to narrow down the specific type of E.coli.
Pathogenic/toxigenic E. coli are of tremendous concern for the fresh produce industry. “Generic” E. coli live in the guts of most mammals; they are typically harmless and may even be beneficial. However, pathogenic/toxigenic E. coli, when ingested by humans, can cause bloody diarrhea and, particularly in the very young and very old, may lead to kidney failure and even death.
Suslow said the new test, which goes by the working name “total pathogenic E. coli,” or TPEC, was tested in a variety of different types of samples: irrigation water, feed lot surface material, manure, compost, soil and produce.
“With all those materials, we had very good effectiveness, but especially with water and produce,” Suslow said.
The research will be presented at the UC Davis Center for Produce Safety Produce Research Symposium on June 23. The Produce Research Symposium is open to the public. A $150 registration fee includes all symposium sessions, breakfast, lunch and an evening reception in the courtyard and gardens of the Robert Mondavi Institute for Wine and Food Science.
For more information or to register for the symposium, see the Center for Produce Safety website at http://cps.ucdavis.edu.

Bagged lettuce was implicated in a recent food safety outbreak.